MetFrag Web
MetFrag Web tool Landing Page call
The new web tool of MetFrag uses a new fragmentation algorithm which allows faster and more efficient processing of the compounds. It has additional fields for setting parameters that depend on your experimental conditions during the MS acquisition. At first the parameters for the candidate database query have to be defined under Database Settings. The candidate database has to be selected which can be queried either by a monoisotopic mass, a molecular formula or comma separated database dependent identifiers. The first has to be defined together with a mass deviation in ppm. A variaty of databases can be selected including PubChem, KEGG, ChEBI and a derivatised version of the KEGG database generated by in silico TMS (trimethylsilylation) and MeOx (methoximation) derivatization.
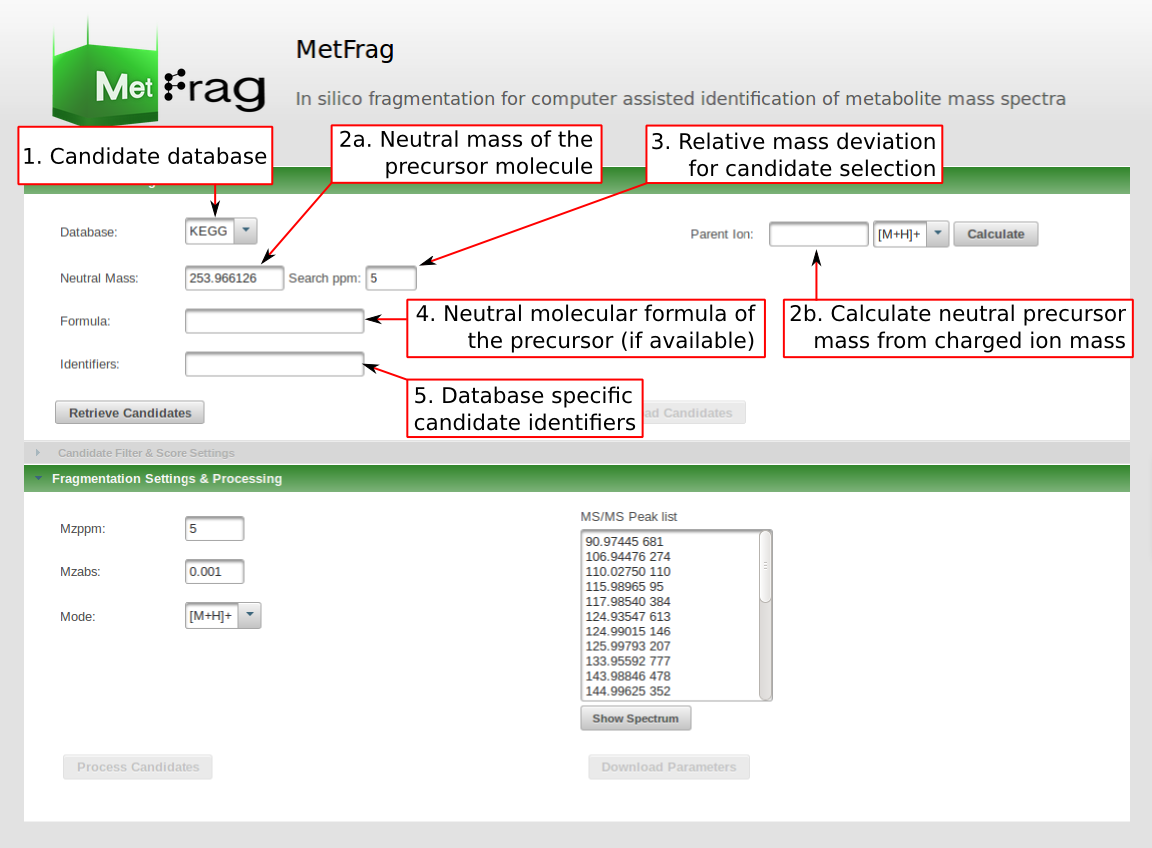
After setting the parameters for the database query the "Retrieve Candidates" button can be pushed to submit the database query.
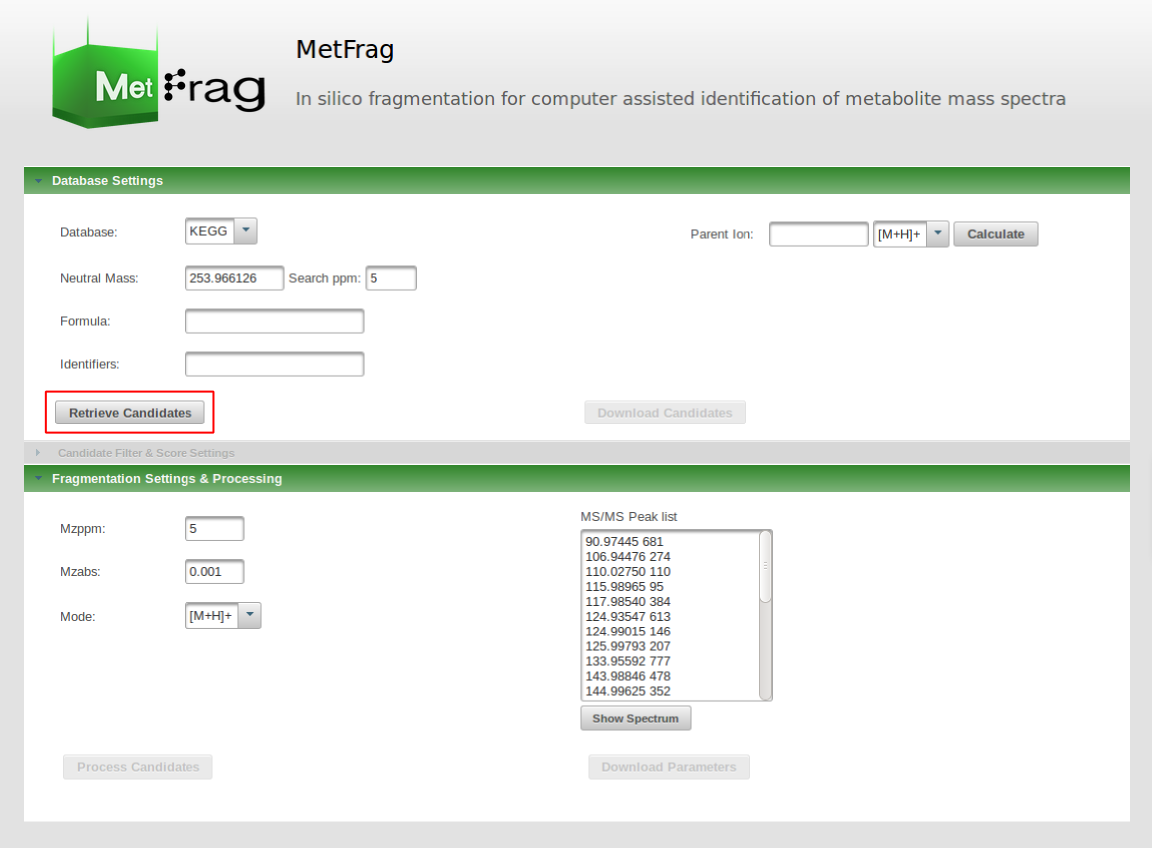
After a short processing time the number of candidates found in the database with the defined filter criteria is displayed. The next step is to define parameters for the MetFrag processing under the Fragmentation Settings & Processing.
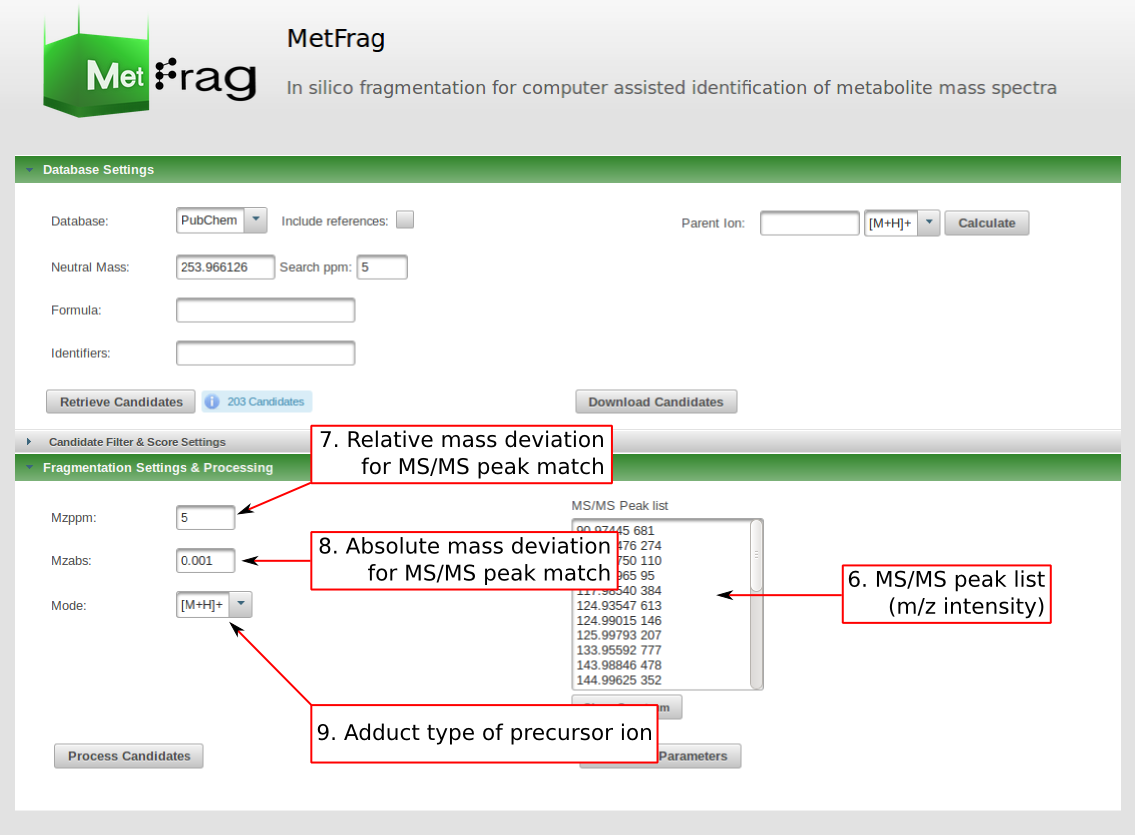
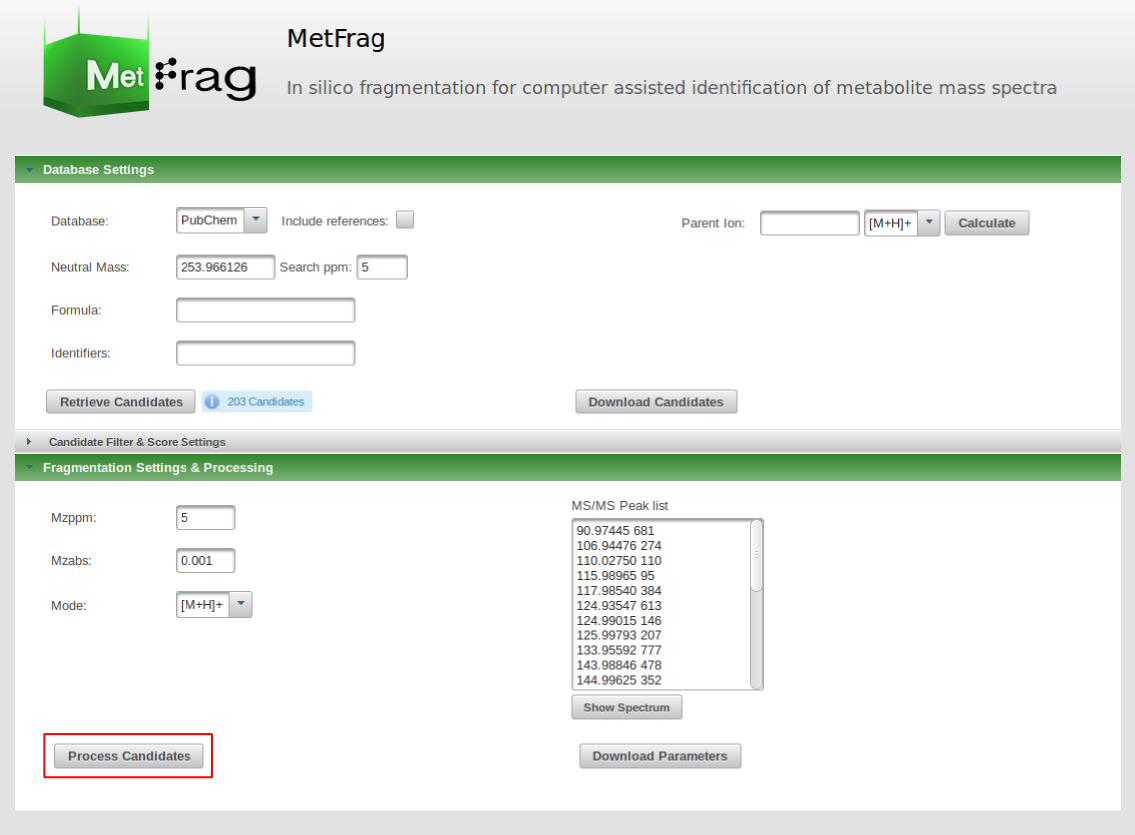
This includes the adduct type of the precursor ion. Based on this fragment massed will be calculated. Additionally, the a relative and absolute mass deviation has to be defined with which in silico generated fragments will be mapped to the experimental data defined in the "MS/MS Peak list" text field. After all parameters are defined correctly, the button "Process Candidates" has to be pushed to start the processing.
MetFrag then starts the in silico fragmentation, the mapping of the fragments to the given mass peaks and the scoring for each retrieved candidates.
MetFrag then starts the in silico fragmentation, the mapping of the fragments to the given mass peaks and the scoring for each retrieved candidates.
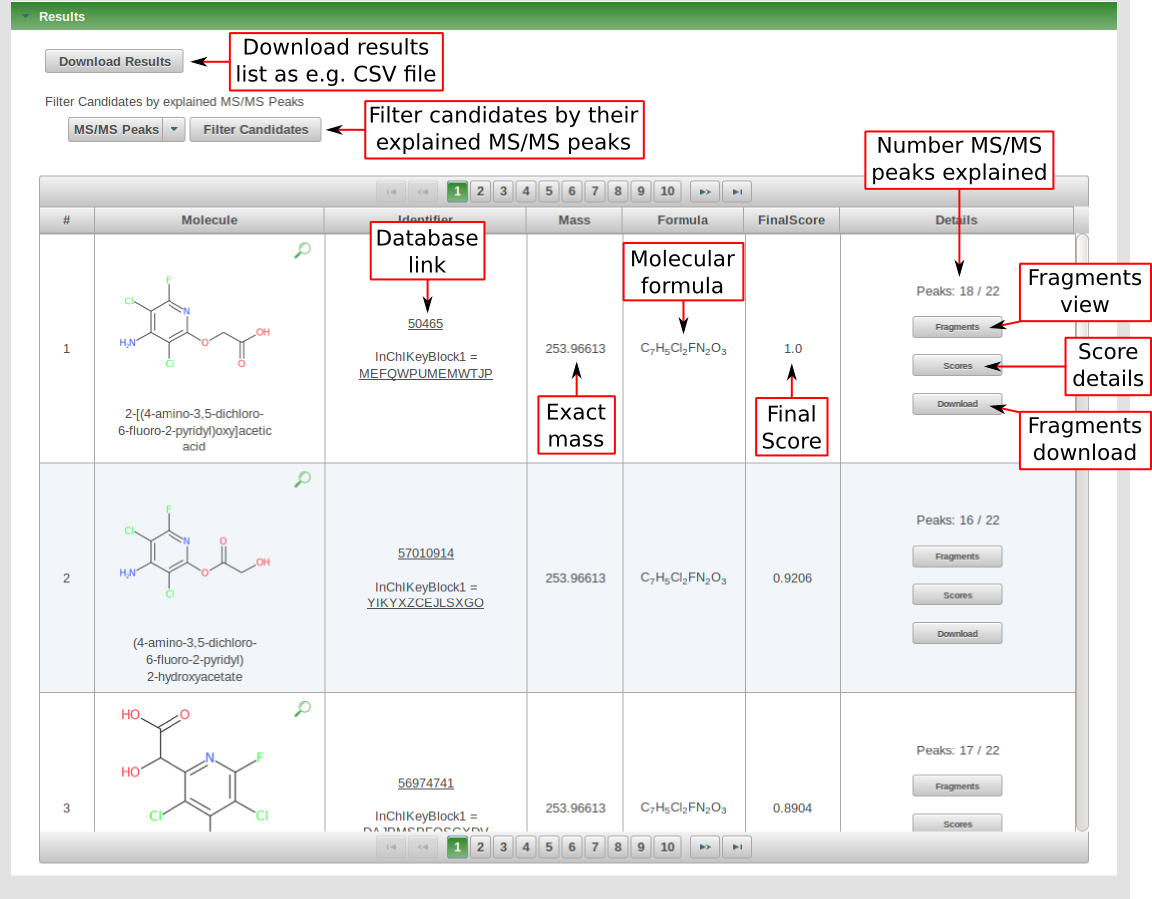
After processing has finished successfully, the score ranked list of candidates will be displayed. Each row in this list includes the candidate image, identifier (database link), exact mass, molecualr formula, score and number of explained together with more details about fragments and scores for each candiate.
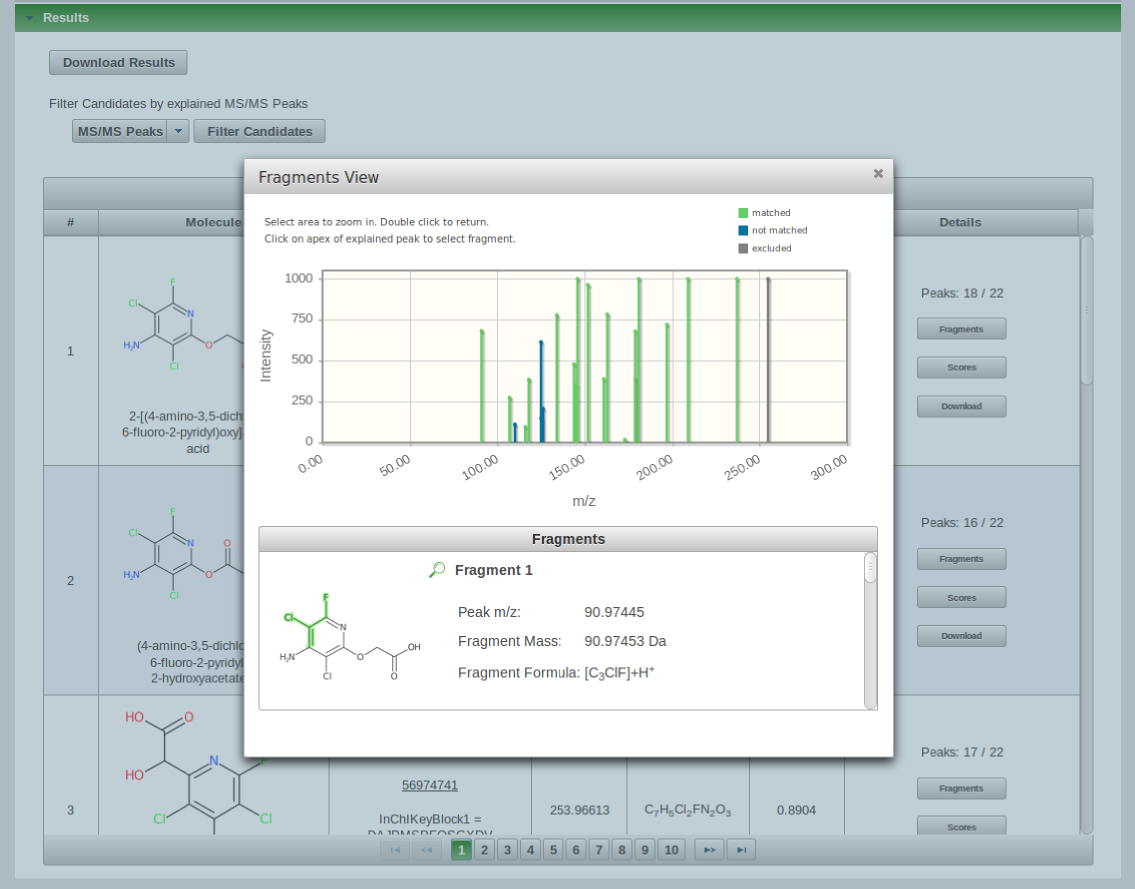
The fragments view shows the assigned fragments for a specific candidate of the list. The substructure from which the fragment was created is highlighted in green color. Each fragment can be viewed by scrolling through the list or by clicking on the apex of an explained peak (green highlighted). With clicking on the peak apex the fragment will be displayed automatically.
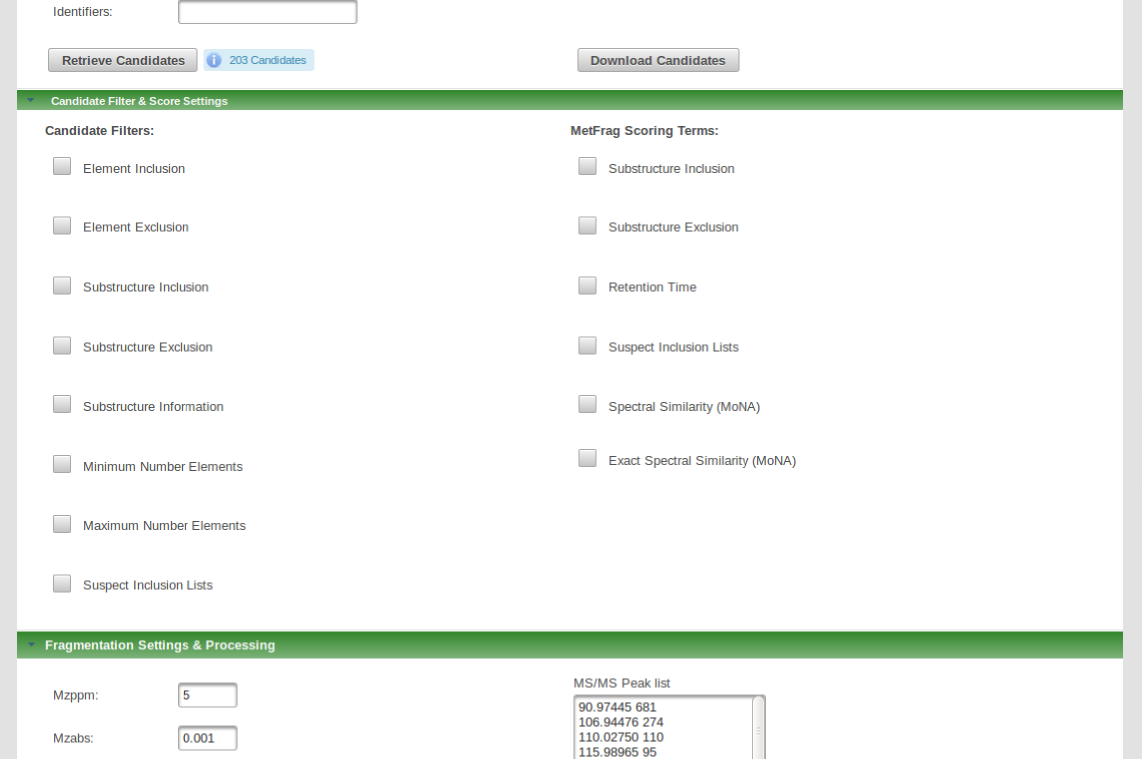
A new feature of the new MetFrag web tool includes the inclusion of additonal candidate filters and scoring terms. This includes candidate filtering by given substructures or by elemental composition or adding several scoring terms by which candidates will be ranked. This includes e.g. scoring by spectral similarity or retention time.
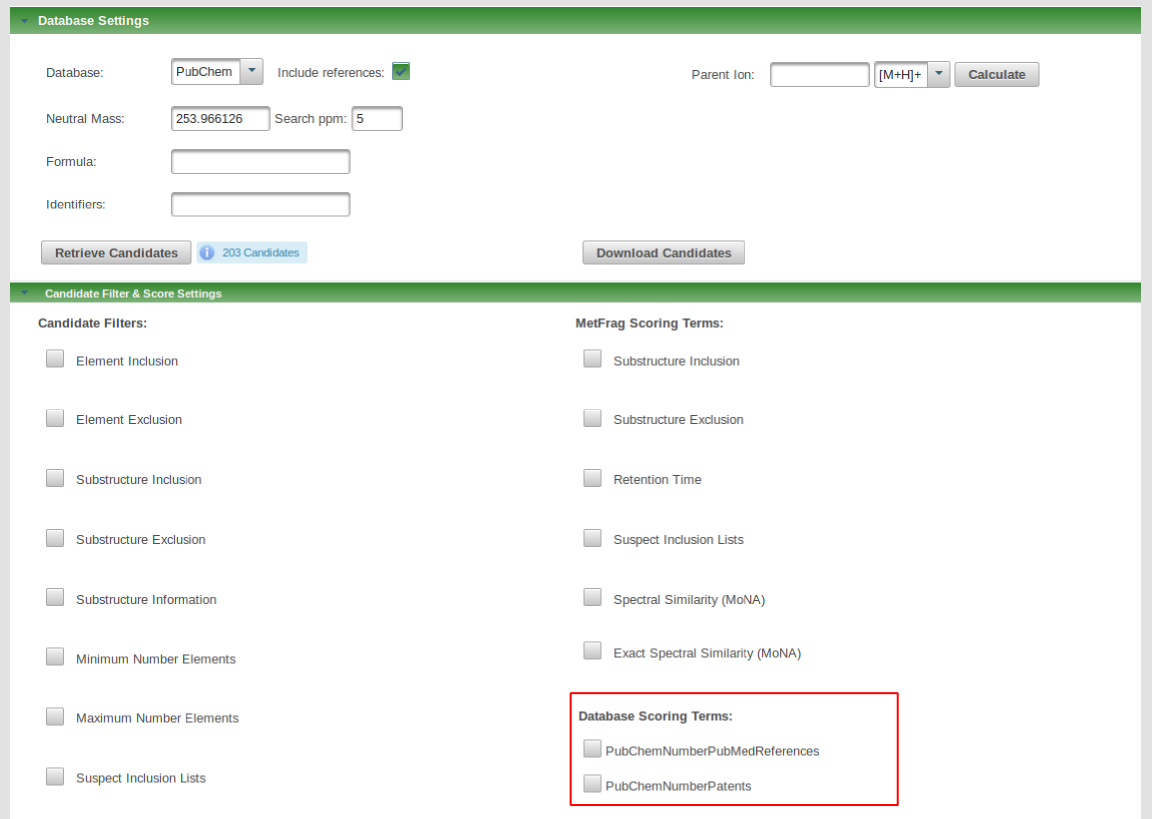
Besides predifined scoring terms, there are also database scoring terms depending on the specified database. PubChem includes PubMed references and patent information that can be used as scoring term. Scoring terms can also be defined by uploaded CSV or SDF files.
MetFrag Landing Page
The Landing page can save the tedious cut+paste experience. All parameters are passed as part of the URL, everything else done in the browser. This allows to link to a query, and even to launch a MetFrag query from another application:




