Discovering regulated Metabolite Families
Khabat Vahabi
Leibniz Institute of Plant BiochemistrySteffen Neumann
Leibniz Institute of Plant Biochemistrysneumann@ipb-halle.de
discoveringregulatedmetabolitefamilies.RmdAbstract
Description of the MetFamily R package
Introduction
Some text about the scenario, data and MTBLS297.
Cool paper [1].
Cool R package: MetFamily
Loading the data
First, we load the data and summarise it
# Load separate files
filePeakMatrixPath <- system.file("extdata/showcase/Metabolite_profile_showcase.txt", package = "MetFamily")
fileSpectra <- system.file("extdata/showcase/MSMS_library_showcase.msp", package = "MetFamily")
fileAnnotation <- system.file("extdata/testdata/canopus/canopus1680.txt", package = "MetFamily")
parameterSet <- parameterSetDefault()
dataList <- projectFromFiles(filePeakMatrixPath,
fileSpectra,
parameterSet = parameterSet,
annot_path =fileAnnotation)## [1] "Parsing MS/MS file..."
## [1] "MS/MS file: Read file"
## [1] "MS/MS file: Parse"
## [1] "MS/MS file: Assemble spectra"
## [1] "MS/MS file: Box"
## [1] "MS/MS file postprocessing"
## [1] "MS/MS file boxing"
## [1] "Parsing MS/MS file ready"
## [1] "Parsing MS1 file..."
## [1] "Parsing MS1 file content..."
## [1] "Precursor deisotoping..."
## [1] "Precursor deisotoping 582 / 5823"
## [1] "Precursor deisotoping 1164 / 5823"
## [1] "Precursor deisotoping 1746 / 5823"
## [1] "Precursor deisotoping 2328 / 5823"
## [1] "Precursor deisotoping 2910 / 5823"
## [1] "Precursor deisotoping 3492 / 5823"
## [1] "Precursor deisotoping 4074 / 5823"
## [1] "Precursor deisotoping 4656 / 5823"
## [1] "Precursor deisotoping 5238 / 5823"
## [1] "Precursor deisotoping 5820 / 5823"
## [1] "Boxing..."
## [1] "Postprocessing matrix..."
## [1] "Building fragment mzFragmentGroups..."
## [1] "Fragment grouping preprocessing..."
## [1] "Fragment grouping preprocessing ready"
## [1] "Fragment grouping"
## [1] "Fragment grouping 54 / 62300"
## [1] "Fragment grouping 31151 / 62300"
##
## [1] "Fragment grouping ready (3.49103379249573s)"
## [1] "Fragment group postprocessing"
## [1] "Fragment group postprocessing: 1394 / 13943"
## [1] "Fragment group postprocessing: 2788 / 13943"
## [1] "Fragment group postprocessing: 4182 / 13943"
## [1] "Fragment group postprocessing: 5576 / 13943"
## [1] "Fragment group postprocessing: 6970 / 13943"
## [1] "Fragment group postprocessing: 8364 / 13943"
## [1] "Fragment group postprocessing: 9758 / 13943"
## [1] "Fragment group postprocessing: 11152 / 13943"
## [1] "Fragment group postprocessing: 12546 / 13943"
## [1] "Fragment group postprocessing: 13940 / 13943"
## [1] "Fragment group postprocessing ready (1.60024857521057s)"
## [1] "Boxing to matrix"
## [1] "Fragment group deisotoping"
## [1] "Fragment group deisotoping 1394 / 13943"
## [1] "Fragment group deisotoping 2788 / 13943"
## [1] "Fragment group deisotoping 4182 / 13943"
## [1] "Fragment group deisotoping 5576 / 13943"
## [1] "Fragment group deisotoping 6970 / 13943"
## [1] "Fragment group deisotoping 8364 / 13943"
## [1] "Fragment group deisotoping 9758 / 13943"
## [1] "Fragment group deisotoping 11152 / 13943"
## [1] "Fragment group deisotoping 12546 / 13943"
## [1] "Fragment group deisotoping 13940 / 13943"
## [1] "Fragment group deisotoping ready (7.99669599533081s)"
## [1] "Fragment group boxing"
## [1] "Building fragment mzFragmentGroups ready"## 'as(<dgCMatrix>, "dgTMatrix")' is deprecated.
## Use 'as(., "TsparseMatrix")' instead.
## See help("Deprecated") and help("Matrix-deprecated").## [1] "Boxing..."
## [1] "Ready"
## [1] "Preprocessing 964 / 2414"
## [1] "Preprocessing 1941 / 2414"
## [1] "Features"
## [1] "Feature postprocessing"
## [1] "Coloring"
## [1] "Coloring init"
## [1] "Coloring naming functions"
## [1] "Coloring gather data"
## [1] "Coloring matrix"
## [1] "Feature annotations"
## [1] "Boxing"
## [1] "Ready"
## [1] "Merging by: featureId and Alignment ID"Filtering data
We can filter the data to remove low quality data points
filterObj <- makeFilterObj(dataList, filter_average = 10000, filter_lfc = 2)
# fileName <- system.file("extdata/testdata/canopus/filterPca_canopus.rds", package = "MetFamily")
# filterObj_file <- readRDS(fileName)
# identical(filterObj_file, filterObj)PCA
PCA on MS1 in Figure @ref(fig:pca).
## [1] "######################################################################################"
## [1] "PCA (Principal Component Analysis)"
## [1] "Analysis: pcaMethods"
PCA of MS1.
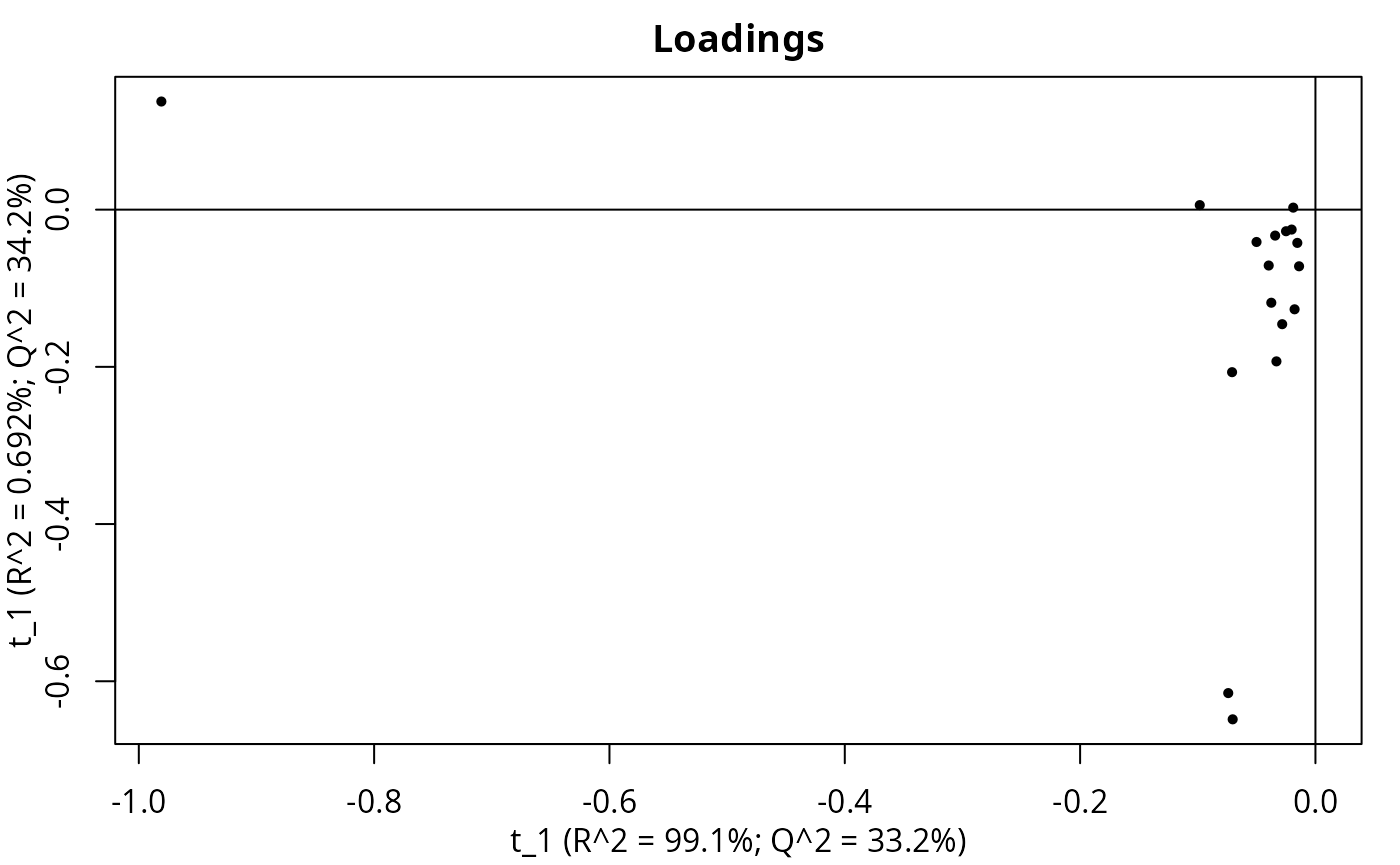
PCA of MS1.
## [1] "######################################################################################"
## [1] "PCA (Principal Component Analysis)"
## [1] "Analysis: pcaMethods"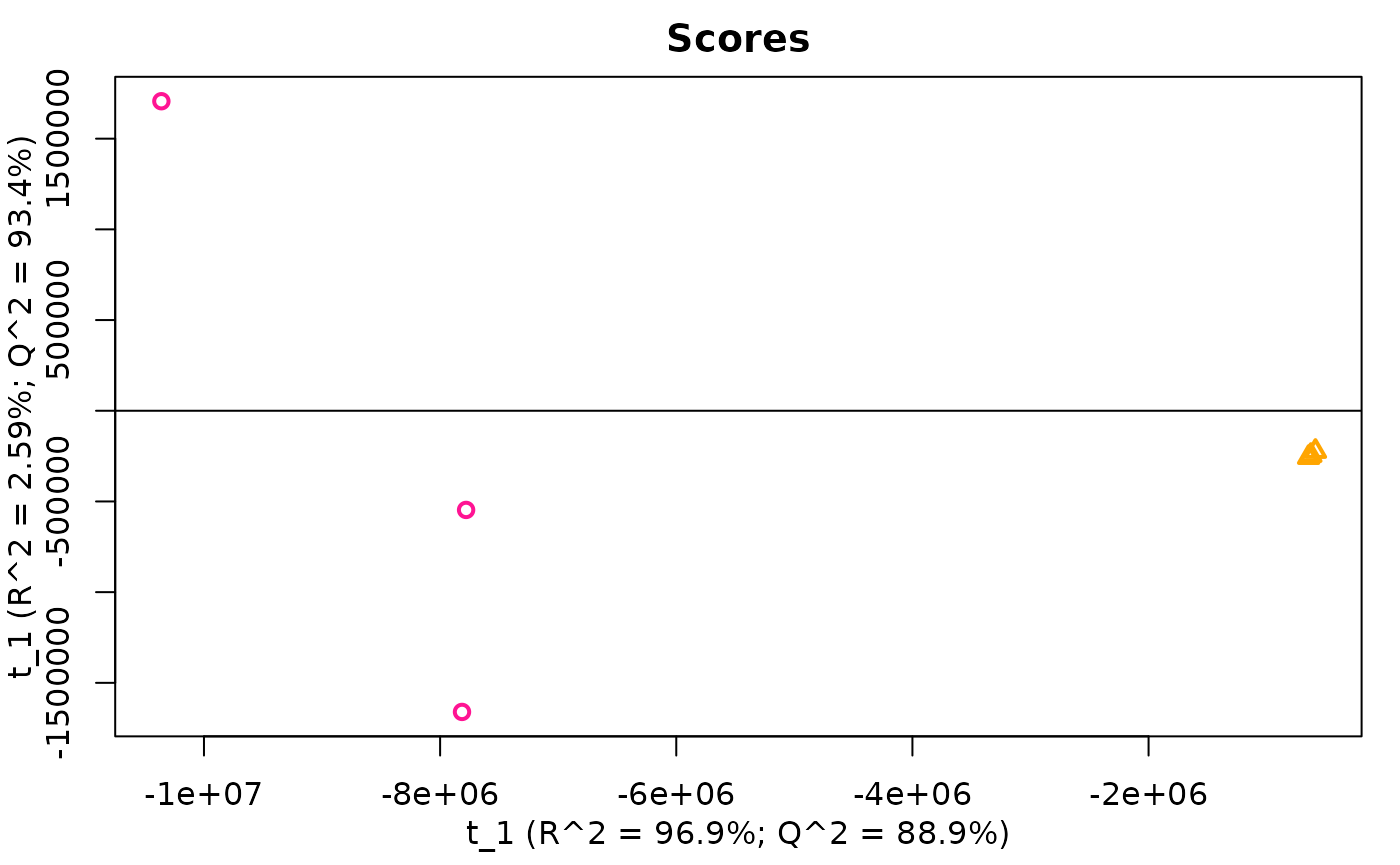
PCA of MS1.
PCA of MS1.
References
1. Treutler H, Tsugawa H, Porzel A, Gorzolka K, Tissier A, Neumann S,
Balcke GU: Discovering
Regulated Metabolite Families in
Untargeted Metabolomics
Studies. Anal Chem 2016.
Appendix
Session info
## R version 4.5.1 (2025-06-13)
## Platform: x86_64-pc-linux-gnu
## Running under: Ubuntu 24.04.2 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
##
## locale:
## [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
## [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
## [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
## [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
##
## time zone: UTC
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] MetFamily_0.99.10 BiocStyle_2.36.0
##
## loaded via a namespace (and not attached):
## [1] DBI_1.2.3 rlang_1.1.6
## [3] magrittr_2.0.3 clue_0.3-66
## [5] MassSpecWavelet_1.74.0 matrixStats_1.5.0
## [7] compiler_4.5.1 systemfonts_1.2.3
## [9] vctrs_0.6.5 reshape2_1.4.4
## [11] stringr_1.5.1 ProtGenerics_1.40.0
## [13] MetaboCoreUtils_1.16.1 pkgconfig_2.0.3
## [15] crayon_1.5.3 fastmap_1.2.0
## [17] XVector_0.48.0 promises_1.3.3
## [19] rmarkdown_2.29 preprocessCore_1.70.0
## [21] UCSC.utils_1.4.0 xcms_4.6.3
## [23] ragg_1.4.0 purrr_1.1.0
## [25] xfun_0.52 MultiAssayExperiment_1.34.0
## [27] cachem_1.1.0 GenomeInfoDb_1.44.1
## [29] jsonlite_2.0.0 progress_1.2.3
## [31] later_1.4.2 DelayedArray_0.34.1
## [33] BiocParallel_1.42.1 prettyunits_1.2.0
## [35] parallel_4.5.1 cluster_2.1.8.1
## [37] R6_2.6.1 bslib_0.9.0
## [39] stringi_1.8.7 RColorBrewer_1.1-3
## [41] limma_3.64.3 GenomicRanges_1.60.0
## [43] jquerylib_0.1.4 iterators_1.0.14
## [45] Rcpp_1.1.0 bookdown_0.43
## [47] SummarizedExperiment_1.38.1 knitr_1.50
## [49] IRanges_2.42.0 BiocBaseUtils_1.10.0
## [51] httpuv_1.6.16 Matrix_1.7-3
## [53] igraph_2.1.4 tidyselect_1.2.1
## [55] abind_1.4-8 yaml_2.3.10
## [57] doParallel_1.0.17 affy_1.86.0
## [59] codetools_0.2-20 lattice_0.22-7
## [61] tibble_3.3.0 plyr_1.8.9
## [63] Biobase_2.68.0 shiny_1.11.1
## [65] evaluate_1.0.4 desc_1.4.3
## [67] Spectra_1.18.2 zip_2.3.3
## [69] affyio_1.78.0 pillar_1.11.0
## [71] BiocManager_1.30.26 MatrixGenerics_1.20.0
## [73] foreach_1.5.2 stats4_4.5.1
## [75] MALDIquant_1.22.3 MSnbase_2.34.1
## [77] plotly_4.11.0 ncdf4_1.24
## [79] generics_0.1.4 hms_1.1.3
## [81] S4Vectors_0.46.0 ggplot2_3.5.2
## [83] scales_1.4.0 MsExperiment_1.10.1
## [85] openxlsx2_1.18 xtable_1.8-4
## [87] glue_1.8.0 MsFeatures_1.16.0
## [89] lazyeval_0.2.2 tools_4.5.1
## [91] mzID_1.46.0 data.table_1.17.8
## [93] vsn_3.76.0 QFeatures_1.18.0
## [95] mzR_2.42.0 XML_3.99-0.18
## [97] fs_1.6.6 grid_4.5.1
## [99] impute_1.82.0 tidyr_1.3.1
## [101] shinyBS_0.61.1 MsCoreUtils_1.20.0
## [103] GenomeInfoDbData_1.2.14 PSMatch_1.12.0
## [105] cli_3.6.5 textshaping_1.0.1
## [107] S4Arrays_1.8.1 viridisLite_0.4.2
## [109] dplyr_1.1.4 AnnotationFilter_1.32.0
## [111] pcaMethods_2.0.0 gtable_0.3.6
## [113] sass_0.4.10 digest_0.6.37
## [115] BiocGenerics_0.54.0 SparseArray_1.8.1
## [117] htmlwidgets_1.6.4 farver_2.1.2
## [119] htmltools_0.5.8.1 pkgdown_2.1.3
## [121] lifecycle_1.0.4 httr_1.4.7
## [123] squash_1.0.9 statmod_1.5.0
## [125] mime_0.13 MASS_7.3-65